Section: Research Program
RNA and protein structures
At the secondary structure level, we contributed novel generic techniques applicable to dynamic programming and statistical sampling, and applied them to design novel efficient algorithms for probing the conformational space. Another originality of our approach is that we cover a wide range of scales for RNA structure representation. For each scale (atomic, sequence, secondary and tertiary structure...) cutting-edge algorithmic strategies and accurate and efficient tools have been developed or are under development. This offers a new view on the complexity of RNA structure and function that will certainly provide valuable insights for biological studies.
Dynamic programming and complexity
Participants : Yann Ponty, Wei Wang, Antoine Soulé, Juraj Michalik.
Common activity with J. Waldispühl (McGill) and A. Denise (Lri ).
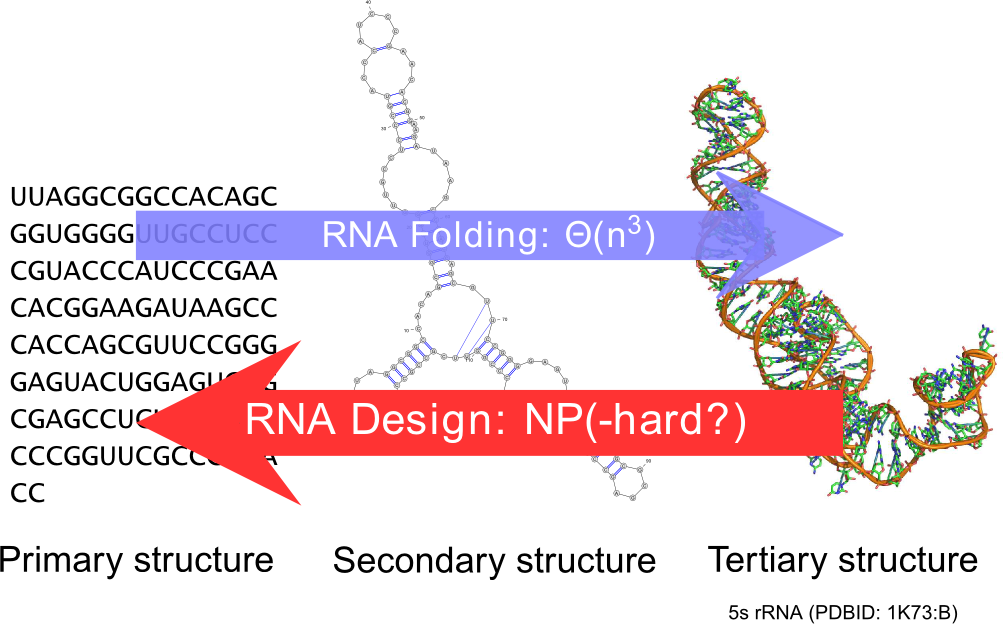
Ever since the seminal work of Zuker and Stiegler, the field of RNA bioinformatics has been characterized by a strong emphasis on the secondary structure. This discrete abstraction of the 3D conformation of RNA has paved the way for a development of quantitative approaches in RNA computational biology, revealing unexpected connections between combinatorics and molecular biology. Using our strong background in enumerative combinatorics, we propose generic and efficient algorithms, both for sampling and counting structures using dynamic programming. These general techniques have been applied to study the sequence-structure relationship [44], the correction of pyrosequencing errors [37], and the efficient detection of multi-stable RNAs (riboswitches) [40], [41].
|
RNA design.
Participants : Alice Héliou, Yann Ponty.
Joint project with A. Denise (sc Lri), J. Waldispühl (McGill), D. Barash (Univ. Ben-Gurion), and C. Chauve (Simon Fraser University).
It is a natural pursue to build on our understanding of the secondary structure to construct artificial RNAs performing predetermined functions, ultimately targeting therapeutic and synthetic biology applications. Towards this goal, a key element is the design of RNA sequences that fold into a predetermined secondary structure, according to established energy models (inverse-folding problem). Quite surprisingly, and despite two decades of studies of the problem, the computational complexity of the inverse-folding problem is currently unknown.
Within our group, we offer a new methodology, based on weighted random generation [24] and multidimensional Boltzmann sampling, for this problem. Initially lifting the constraint of folding back into the target structure, we explored the random generation of sequences that are compatible with the target, using a probability distribution which favors exponentially sequences of high affinity towards the target. A simple posterior rejection step selects sequences that effectively fold back into the latter, resulting in a global sampling pipeline that showed comparable performances to its competitors based on local search [31].
Towards 3D modeling of large molecules
Participants : Yann Ponty, Afaf Saaidi, Mireille Régnier, Amélie Héliou.
Joint projects with A. Denise (Lri) , D. Barth (Versailles), J. Cohen (Paris-Sud), B. Sargueil (Paris V) and Jérome Waldispühl (McGill).
The modeling of large RNA 3D structures, that is predicting the three-dimensional structure of a given RNA sequence, relies on two complementary approaches. The approach by homology is used when the structure of a sequence homologous to the sequence of interest has already been resolved experimentally. The main problem then is to calculate an alignment between the known structure and the sequence. The ab initio approach is required when no homologous structure is known for the sequence of interest (or for some parts of it). We contribute methods inspired by both of these settings directions.
Modeling tasks can also be greatly helped by the availability of experimental data. However, high-resolution techniques such as crystallography or RMN, are notoriously costly in term of time and ressources, leading to the current gap between the amount of available sequences and structural data. As part of a colloboration with B. Sargueil's lab (Faculté de pharmacie, Paris V) funded by the Fondation pour la Recherche medical, we strive to propose a new paradigm for the analysis data produced using a new experimental technique, called Shape analysis (Selective 2'-Hydroxyl Acylation analyzed by Primer Extension). This experimental setup produces an accessibility profile associated with the different positions of an RNA, the shadow of an RNA. As part of A. Saaidi's PhD, we currently design new algorithmic strategies to infer the secondary structure of RNA from multiple Shape experiments performed by experimentalists at Paris V. Those are obtained on mutants, and will be coupled with a fragment-based 3D modeling strategy developed by our partners at McGill.